- Mazarine Desplanque, supervisors : Véronique Bernard and Nicolas Pietrancosta.
Acetylcholine/Glutamate co-transmission in the striatal network. Anatomical and functional heterogeneity of synaptic vesicles in cholinergic interneurons - Konstantin Volzhenin, supervisors : Alessandra Carbone and Lucie Bittner.
Deep learning to predict metabolic protein-protein interaction networks for environmental microbial communities - Zoheir Ziriat, supervisors : Laure Teysset, Clément Carré and Christophe Antoniewski. Study of piRNA clusters: loci involved in genome stability
- Cécile Vincent, supervisors : Elie Wandersman and Vincent Noireaux. A Biomimetic Approach of Cell-Cell Communication
Interface Biology and Chemistry
Meeting with Mazarine Desplanque

My scientific background
As a student of the pharmacy-science program of the “Ecole de l’Inserm”, I have been aspiring for years to become a pharmacist as well as a researcher. Early I had the opportunity to work with chemists, biologists, pharmacists and physicians and these experiences convinced me of the importance of preserving a transversal thinking between fundamental science and medical knowledge. Therefore, I felt it was essential to undertake a PhD thesis in order to effectively work towards this endeavor of mine.
What attracts me to the PhD experience
I am truly looking forward to achieving a PhD experience as my different internships gave me the desire to conduct my own experiments and assume a greater role in science. I am also enthusiastic about exchanging with the two laboratories. Furthermore, undertaking a PhD at this stage of my studies, before my pharmacy externship, will then allow me to finish my pharmaceutical studies with a new vision.
My future research
The striatum plays a central role in the regulation of reward and habits-guided behaviors as well in locomotor activity. Striatal activity is regulated by cholinergic interneurons (CINs) that display unique molecular characteristics: they express vesicular transporter for acetylcholine (VAChT) and for glutamate (VGLUT) and consequently regulate the striatal network with both ACh and Glu. However, the specific morphological and functional characteristics of ACh/Glu co-transmission are still poorly understood. Our hypothesis is that CINs contain three pools of synaptic vesicles expressing either VAChT, VGLUT3 or both releasing ACh, Glu or both. To explore this hypothesis, we will use a multidisciplinary approach combining super resolution microscopy, new chemical tools (fluorescent sensors to locate VGLUT) and optogenetic stimulations. This project will lead to a deeply understanding of the striatum and could open the way to new treatments for addiction or Parkinson disease. It will not only bring knowledge progress, it will also contribute to the development of pivotal tools such as fluorescent VGLUT ligands, bridging neurosciences and pharmacological research.
Figure
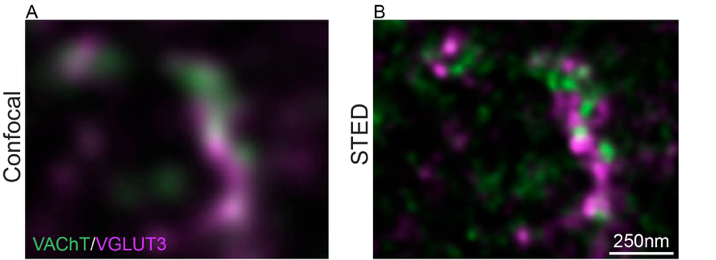
Find out more:
[1] El Mestikawy, Salah et al. “From glutamate co-release to vesicular synergy: vesicular glutamate transporters.” Nature reviews. Neuroscience vol. 12,4 (2011): 204-16. doi:10.1038/nrn2969
[2] Gubernator, N. G. et al. Fluorescent false neurotransmitters visualize dopamine release from individual presynaptic terminals. Science 324, 1441-1444, doi:10.1126/science.1172278 (2009).
[3] S Ren, J. et al. Habenula “Cholinergic” Neurons Corelease Glutamate and Acetylcholine and Activate Postsynaptic Neurons via Distinct Transmission Modes. Neuron 69, 445-452 (2011), doi:10.1016/j.neuron.2010.12.038
Contact
Mazarine DESPLANQUE
Email: mazarinedesplanque1999@gmail.com
- Team “Neuropharmacology of VGLUTs”
Neurosciences Paris Seine (NPS) – CNRS UMR 8246 – INSERM U1130- Sorbonne Université
Institut Biologie Paris-Seine
7-9 quai Saint Bernard – Bât B 4ème étage
75 252 Paris CEDEX, France
https://www.ibps.sorbonne-universite.fr/fr/Recherche/umr-8246/systemes-glutamatergiques-normaux-et-pathologiques - Team “Biomolecules: Analysis, Molecular and Cellular Interactions”
Laboratoire des BioMolécules (LBM), UMR 7203, CNRS – ENS – Sorbonne Université
4 place Jussieu – Tour 23-33
Paris 75005, France
https://lbm.cnrs.fr/
Interface Biology and Maths/Modeling/Computer science
Meeting with Konstantin Volzhenin

My scientific background
I have a general background in Physics and Mathematics, however, during my Master studies, I became interested in Artificial Intelligence and its applications in Biology in particular. I have worked in both industrial and academic research, applying my knowledge to different topics, such as Neuroscience or Robotics. My current project in Metagenomics tackles very interesting and exciting questions in a field that can greatly benefit from new AI tools.
What attracts me to the PhD experience
I think that PhD research provides unique opportunities to set a creative atmosphere. This allows implementing of various interesting ideas and finding nontrivial solutions. Moreover, I find it very exciting to work on unusual and difficult problems on a regular basis and I believe that academic research is a perfect place for this.
My future research
My work will be dedicated to developing a new computational approach to reconstruct protein-protein interaction (PPI) networks for metagenomic samples. I will create a Deep Learning model that will have to deal with a specific classification task: it will predict which proteins from a specific microbial community can interact with each other and it will estimate the likelihood of such interactions. This work has never been done before for a group of organisms thus it can serve as an important tool to study Ecosystems Biology on the smallest, molecular scale. I am very excited by the interdisciplinary nature of this project which combines state-of-the-art AI research with large-scale biological datasets in order to shed some light on incredibly complex interactions in the living world.
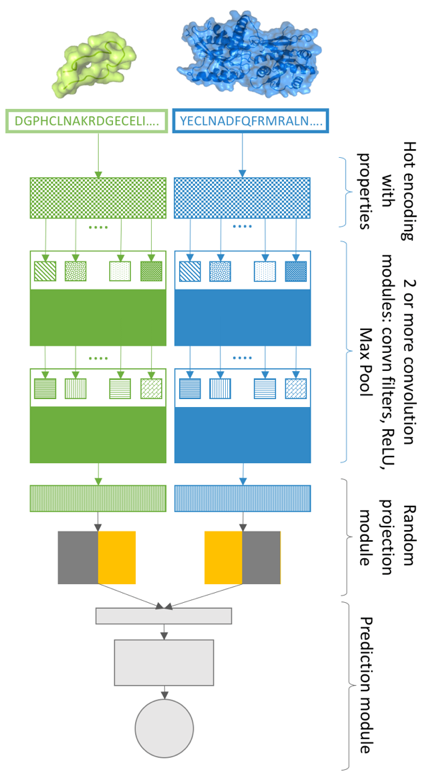
Figure
Find out more
[1] Vicedomini R, Bouly JP, Laine E, Falciatore A, Carbone A. Multiple probabilistic models extract features from protein sequence data and resolve functional diversity of very different protein families. bioRxiv. 2021 Jan 1;717249. https://www.biorxiv.org/content/10.1101/717249v2
[2] Ugarte A, Vicedomini R, Bernardes J, Carbone A. A multi-source domain annotation pipeline for quantitative metagenomic and metatranscriptomic functional profiling. Microbiome. 2018 Aug 28;6(1):149. https://doi.org/10.1186/s40168-018-0532-2
Contact
Konstantin VOLZHENIN
Email: Konstantin_v_v@outlook.com
- Team “Analytical Genomics”
Laboratory of Computational and Quantitative Biology (LCQB) UMR 7238 CNRS – Sorbonne University
Institut de Biologie Paris-Seine
7-9 Quai Saint Bernard, Bâtiment C, 4ème étage
75005 Paris, France
https://www.ibps.sorbonne-universite.fr/en/research/computational-and-quantitative-biology/analatical-genomics - Team “Atelier de bio-informatique“
Institut de Systématique, Evolution, Biodiversité (SYEB) UMR 7205 CNRS – EPHE – Sorbonne University
Muséum National d’Histoire Naturelle
45 rue Buffon, Bâtiment d’Entomologie
75005 Paris, France
https://isyeb.mnhn.fr/fr/atelier-de-bio-informatique-384
Interface Biology and Maths/Modeling/Computer science
Meeting with Zoheir Ziriat

My scientific background
I have always been interested in epigenetic mechanisms and all the fascinating and incredible ways they allow the regulation of gene expression. During my Master on Biochemistry and Genetics of RNA at Sorbonne University and internships, I’ve met many researchers and PhD students working on these topics, all with very different techniques and approaches. These meetings confirmed my interest in the study of epigenetic and motivated me to do a PhD in order to deepen our understanding of this research field.
What attracts me to the PhD experience
For me, a PhD is like diving into a maze and trying, using different techniques and exploring different directions, to find our way to its center and the scientific truth that lies within. I am very excited to undertake this long-term exploration of a subject that fascinates me, and looking forward to learning new techniques, analyzing my results, interacting with others and managing this project.
My future research
At the beginning, there were Transposable Elements (TEs). These are DNA sequences capable of moving and inserting themselves elsewhere in a genome. They represent a huge part of eukaryotic genomes, and thus, could be deleterious. To control TEs, evolution has developed a mechanism based on small non-coding RNAs called piRNAs. These are transcribed from heterochromatic loci composed of fragments of TEs: the piRNA clusters, and we know very little about these genomic regions.
My PhD project aims to decipher the mechanisms behind the evolution and the regulation of piRNA clusters in order to understand how these loci react to the insertion of a new TE and how their transcripts are recognized as piRNA precursors. To do so, I will be using Nanopore 3rd generation sequencer to study piRNA clusters transcripts, and different types of informatics algorithms to try to detect potential chemical RNA modifications in their sequence that could act as tags.
Figure
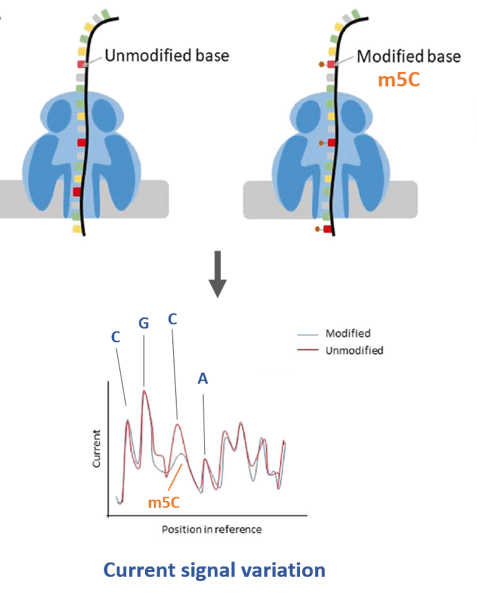
Find out more
[1] Czech, Benjamin, et Gregory J. Hannon. « One Loop to Rule Them All: The Ping-Pong Cycle and PiRNA-Guided Silencing ». Trends in Biochemical Sciences 41, no 4 (2016): 324-37. https://doi.org/10.1016/j.tibs.2015.12.008
[2] Liu, Huanle, Oguzhan Begik, Morghan C. Lucas, Jose Miguel Ramirez, Christopher E. Mason, David Wiener, Schraga Schwartz, John S. Mattick, Martin A. Smith, et Eva Maria Novoa. « Accurate Detection of m 6 A RNA Modifications in Native RNA Sequences ». Nature Communications 10, no 1 (2019): 4079. https://doi.org/10.1038/s41467-019-11713-9
Contact
Zoheir ZIRIAT
Email: zoheir.ziriat@sorbonne-universite.fr
- Team “TErBio: Transgenerational epigenetics & small RNA biology”
Developmental Biology Laboratory (LBD) UMR 7622, CNRS, Sorbonne University
Institut de Biologie Paris-Seine
9 quai Saint Bernard, case courrier 24
75 252 PARIS CEDEX, France
https://www.ibps.sorbonne-universite.fr/en/research/developmental-biology-laboratory/transgenerational-epigenetics-small-rna-biology - ARTbio bioinformatics analyses platform, FR3631 CNRS – Sorbonne University
Institut de Biologie Paris-Seine
9 quai Saint Bernard
75 252 PARIS CEDEX, France
https://www.artbio.fr/
Interface Biology and Physics
Meeting with Cécile Vincent

My scientific background
After a bachelor in Physics, I studied fundamental Physics and then during my second year of Master I specialized in fluid mechanics. I have always been interested in the application of Physics to Biology. Indeed, during my Master I did an internship at the interface of Physics and Biology and I enjoyed working on an interdisciplinary project. That’s why I wanted to do a PhD in Biophysics.
What attracts me to the PhD experience
Since the beginning of my studies, I knew that I wanted to do a PhD. I think that doing a PhD is the perfect opportunity to develop and improve knowledge in science. My thesis project will allow me to build an experimental setup and learn new scientific techniques. I find it very stimulating to carry out a project requiring high-level science.
My future research
Cell-cell communication participates in regulating and synchronizing cellular functions. In tissues, the transport of ions and molecules from a cell to another occurs in particular through protein nanometric pores across the cell membrane. Such nanopores can be inert (simple nano-holes) or mechanosensitive with an ionic/molecular permeability that depends on the stress acting on the membrane. This project proposes a biomimetic approach of cell-cell communication in tissues. Tissues will be mimicked with 2D arrays of aqueous droplets connected by lipid membranes decorated with transmembrane proteins, inert or mechanosensitive. Mechanosensitive proteins will be synthetized directly within the droplets using cell free Transcription Translation reactions. For inert networks, we will probe with epifluorescence microscopy how the diffusion of Ca2+ ions depends on the network topology and pore concentration. For mechanosensitive networks, we will probe with a rheo-microscope how controlled deformations of the network affect the transport properties. Our results will be modeled using random walks in nanoporous media, in which the opening gate probability depends on the local stress.
Figure

Find out more
Diffusion through Nanopores in Connected Lipid Bilayer Networks, Physical Review Letters, M. Valet, L.-L. Pontani, R. Voituriez, E. Wandersman, A. Prevost. 123, 088101, 2019. DOI: https://doi.org/10.1103/PhysRevLett.123.088101
Contact
Cécile VINCENT
Email : cecile.vincent@sorbonne-universite.fr
- Team “Mechanics of integrated and artificial biological systems”
Laboratoire Jean Perrin (LJP) UMR 8237 CNRS – Sorbonne University
Institut de Biologie Paris-Seine – 4 place Jussieu, Tour 32-33 – 75005 Paris, France - Team “Physics and Nanotechnology”
Minneapolis, MN 55455 – USA, University of Minnesota
