4 Interdisciplinary collaborative projects have received an i-Bio grant in 2022 :
- Breau Marie (UMR7622-IBPS)
Bonnet Isabelle (UMR168-Institut Curie)
Click here to discover the project -> Deciphering the biomechanical mechanisms driving the expansion of the olfactory orifice in zebrafish
- Cauli Bruno (UMR8246-IBPS)
Papagiakoumou Eirini (UMR7210-Institut de la Vision)
Click here to discover the project -> Probing Metabolic Tuning of Neurovascular Coupling by multimodal imaging and holographic optogenetics (MetaTuNe)
- Nizak Clément (UMR8237-IBPS)
Weigt Martin (UMR7238-IBPS)
Click here to discover the project -> Statistical Design of Serine Protease Substrate Specificity
- Paradis Anne-Lise (UMR8246-IBPS)
Navarro Vincent (UMR7225-Brain Institute)
Click here to discover the project -> Development of a new computerized experimentation tool to study visual exploration and space coding in epileptic patients
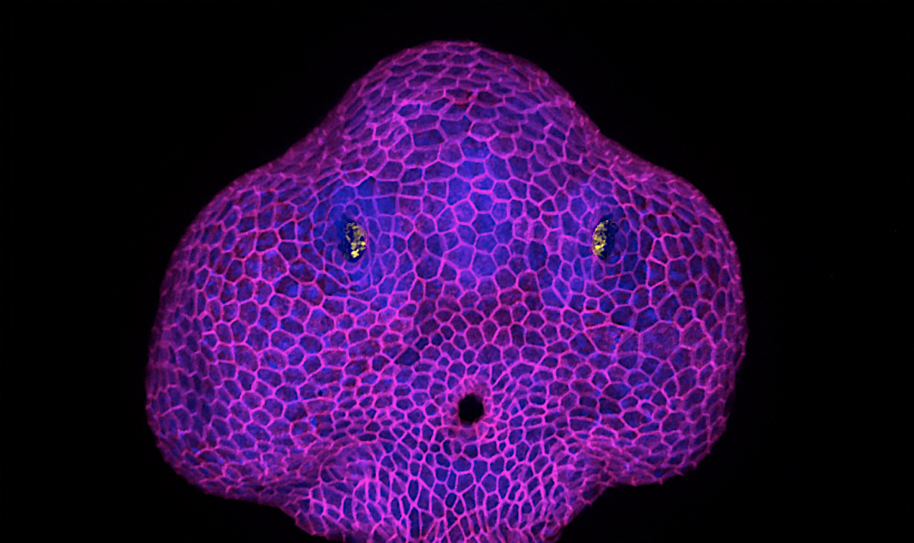
Deciphering the biomechanical mechanisms driving the expansion of the olfactory orifice in zebrafish
Epithelia can lose their integrity to create physiological openings that persist and grow over time.We will investigate the underlying biophysical mechanisms, a poorly explored question in the field of epithelial morphogenesis. We will study the zebrafish olfactory organ, in which a hole (the future nostril) forms in the skin epithelium to expose the olfactory neurons to odor cues. Our previous results suggest that the neurons pull on the skin cells to trigger the opening of the epithelium. Here, we will analyse how the growth of the orifice occurs. Preliminary data show that the hole grows in a region of the skin where cell packing differs from what is further away, and above a rosette of olfactory neurons that seems to steer the process by changing its shape. We will test the role of the neuronal rosette rearrangement and of the skin remodelling around the orifice. We will combine live imaging with biophysical tools such as laser ablation and optogenetics to map and perturb forces.
Marie BREAU
Developmental Biology Laboratory (UMR7622), Institut Biologie Paris-Seine (IBPS)
Team : Mechanics of Neuronal Development
Email : marie.breau@upmc.fr
Isabelle BONNET
Physical Chemistry Curie (UMR 168), Institut Curie
Team : Biology-Inspired Physics at Mesoscales
Email : isabelle.bonnet@curie.fr
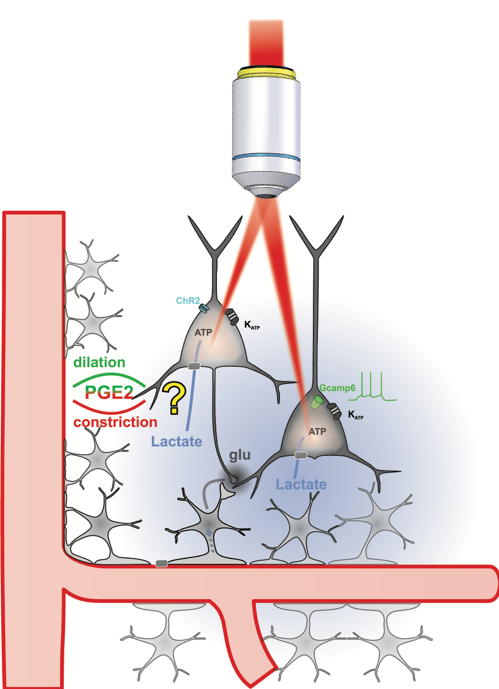
Probing Metabolic Tuning of Neurovascular Coupling by multimodal imaging and holographic optogenetics (MetaTuNe)
Local regulation of cerebral blood flow and metabolism through neuronal activity is essential for brain function. Neurovascular and neurometabolic coupling ensure cerebral glucose homeostasis and lead to a rise in extracellular lactate, the excess of which can be deleterious. We recently showed that lactate is an energy substrate for cortical neurons enhancing their activity [1], and that vasodilating neurons become vasoconstrictors at high-level spiking activities. This frequency-polarized vasoconstriction might restrict excessive lactate supply to maintain brain activity balance.
To explore this hypothesis, we will determine how lactate shapes neuronal activity and neurovascular coupling using integrative ex vivo and in vivo imaging combined with cutting-edge multiphoton holographic optogenetics[2]. If successful, this project will deepen the understanding of how metabolic states influence neurovascular functions and help to identify new therapeutic targets for neurological disorders.
[1] Karagiannis A, Gallopin T, Lacroix A, Plaisier F, Piquet J, Geoffroy H, Hepp R, Naudé J, Le Gac B, Egger R, Lambolez B, Li D, Rossier J, Staiger JF, Imamura H, Seino S, Roeper J, Cauli B (2021). Lactate is an energy substrate for rodent cortical neurons and enhances their firing activity. Elife 10, e71424; doi: 10.7554/eLife.71424.
[2] Chen IW, Ronzitti E, Lee BR, Daigle TL, Dalkara D, Zeng H, Emiliani V, Papagiakoumou E (2019). In Vivo Submillisecond Two-Photon Optogenetics with Temporally Focused Patterned Light. J. Neurosci. 39, 3484-3497; doi: 10.1523/JNEUROSCI.1785-18.2018.
Bruno CAULI
Neuroscience Paris Seine (UMR 8246 CNRS, U 1130 Inserm), Institut Biologie Paris-Seine (IBPS), Sorbonne Université.
Team: Synaptic & Neuroenergetic Networks
Email: bruno.cauli@upmc.fr
Eirini PAPAGIAKOUMOU
Photonics Department (UMR 7210 CNRS, UMR S968 Inserm) Institut de la Vision, Sorbonne Université.
Team: Wavefront Engineering Microscopy
Email: eirini.papagiakoumou@inserm.fr
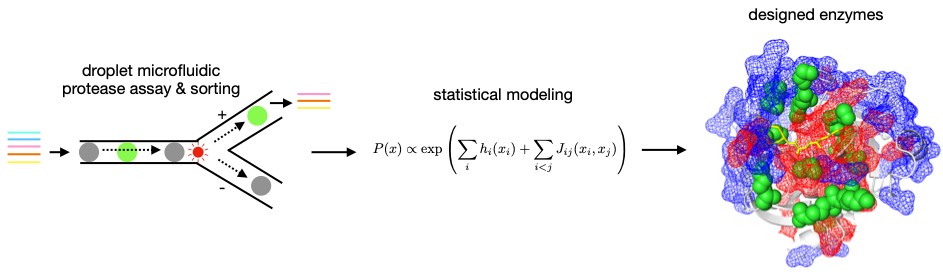
Statistical Design of Serine Protease Substrate Specificity
We aim at understanding the relation between enzyme sequence and substrate specificity using serine proteases as a model system. These enzymes have been extensively characterized, but genetically engineering their substrate specificity has proven challenging [1]. Our new interdisciplinary approach considers this problem in protein sequence space with no required input in terms of protein structure and catalytic mechanism. We generate high throughput experimental data to derive interpretable, statistical physics-inspired models of the relation between enzyme sequence and catalytic activity towards specific substrates. For this, we measure the catalytic activity of hundreds of proteases towards several peptide substrates in a single experiment based on droplet microfluidics [2]. The experimental datasets will feed our statistical models, which we will build and train to design sequences of synthetic proteases with arbitrary substrate specificity that we will validate experimentally.
[1] L. Hedstrom, Serine protease mechanism and specificity. Chemical reviews. 102, 4501–4524 (2002).
[2] J. J. Agresti, E. Antipov, A. R. Abate, K. Ahn, A. C. Rowat, J.-C. Baret, M. Marquez, A. M. Klibanov, A. D. Griffiths, D. A. Weitz, Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proceedings of the National Academy of Sciences of the United States of America. 107, 4004–4009 (2010).
Clément NIZAK
Jean Perrin Laboratory (UMR 8237), Institut Biologie Paris-Seine (IBPS)
Theme : Micro-organisms biophysics
Email : clement.nizak@upmc.fr
Martin WEIGT
Computational and Quantitative Biology (UMR 7238), Institut Biologie Paris-Seine (IBPS)
Team: Statistical Genomics and Biological Physics
Email : martin.weigt@upmc.fr
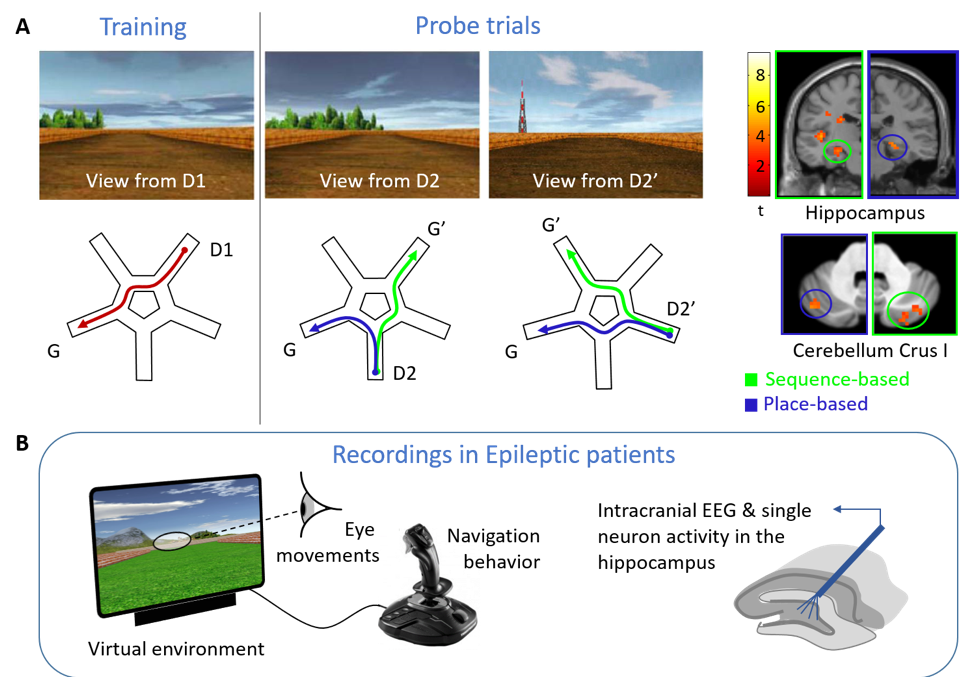
Development of a new computerized experimentation tool to study visual exploration and space coding in epileptic patients
Even in a familiar city, we may get lost and feel disoriented when unexpectedly stumbling upon a known place. We hypothesize that a switch then occurs between a ‘memory-driven’ mode, in which the hippocampus updates the expected location based on monitoring self-motion sequences, and a ‘sensory-driven’ mode, in which the hippocampus retrieves external landmarks to (re)anchor internal maps to actual places [1]. To determine whether gaze may trigger switches in behavior and hippocampal activity, we will develop a computerized tool providing a controlled virtual environment for navigation, interfaced with eye-tracking and deep EEG recording. To identify changes at the neuron coding level, we will record hippocampal activity in epileptic patients using deep brain electrodes implanted for pre-surgical evaluation [2], and assess whether a navigation strategy known to involve hippo-cerebellar connectivity may influence epilepsy-related signals in the hippocampus.
[1] Iglói K, Doeller CF, Paradis AL, Benchenane K, Berthoz A, Burgess N, Rondi-Reig L (2015) Interaction between hippocampus and cerebellum crus i in sequence-based but not place-based navigation. Cereb Cortex 25 (11): 4146–4154.
[2] Alvarado-Rojas C, Lehongre K, Bagdasaryan J, Bragin A, Staba R, Engel J, Navarro V, Le Van Quyen M (2013) Single-unit activities during epileptic discharges in the human hippocampal formation. Front Comput Neurosci 7 (October): 140.
Anne-Lise PARADIS
Neuroscience Laboratory (UMR 8246), Institut Biologie Paris-Seine (IBPS)
Team : Cerebellum, Navigation And Memory (CEZAME)
Email : anne-lise.paradis@upmc.fr
Vincent NAVARRO
Paris Brain Institute (Inserm U 1127, CNRS UMR 7225)
Team: Dynamics of Epileptic Networks and Neuronal Excitability
Email : vincent.navarro@aphp.fr
